Complete benchmark experiment to compare different learning algorithms across one or more tasks w.r.t. a given resampling strategy. Experiments are paired, meaning always the same training / test sets are used for the different learners. Furthermore, you can of course pass “enhanced” learners via wrappers, e.g., a learner can be automatically tuned using makeTuneWrapper.
benchmark( learners, tasks, resamplings, measures, keep.pred = TRUE, keep.extract = FALSE, models = FALSE, show.info = getMlrOption("show.info") )
Arguments
| learners | (list of Learner | character) |
|---|---|
| tasks | list of Task |
| resamplings | (list of ResampleDesc | ResampleInstance) |
| measures | (list of Measure) |
| keep.pred | ( |
| keep.extract | ( |
| models | ( |
| show.info | ( |
Value
See also
Other benchmark:
BenchmarkResult,
batchmark(),
convertBMRToRankMatrix(),
friedmanPostHocTestBMR(),
friedmanTestBMR(),
generateCritDifferencesData(),
getBMRAggrPerformances(),
getBMRFeatSelResults(),
getBMRFilteredFeatures(),
getBMRLearnerIds(),
getBMRLearnerShortNames(),
getBMRLearners(),
getBMRMeasureIds(),
getBMRMeasures(),
getBMRModels(),
getBMRPerformances(),
getBMRPredictions(),
getBMRTaskDescs(),
getBMRTaskIds(),
getBMRTuneResults(),
plotBMRBoxplots(),
plotBMRRanksAsBarChart(),
plotBMRSummary(),
plotCritDifferences(),
reduceBatchmarkResults()
Examples
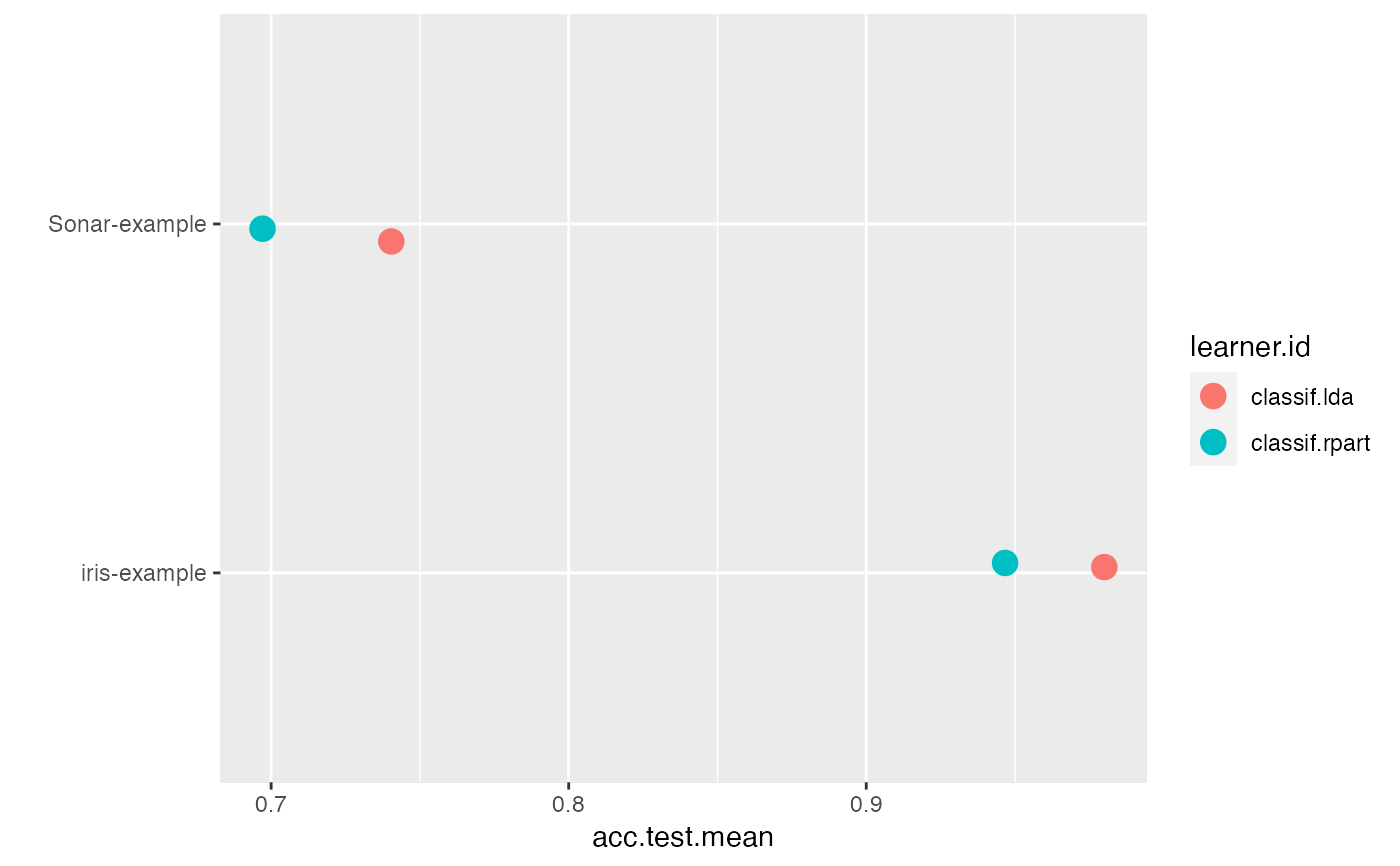
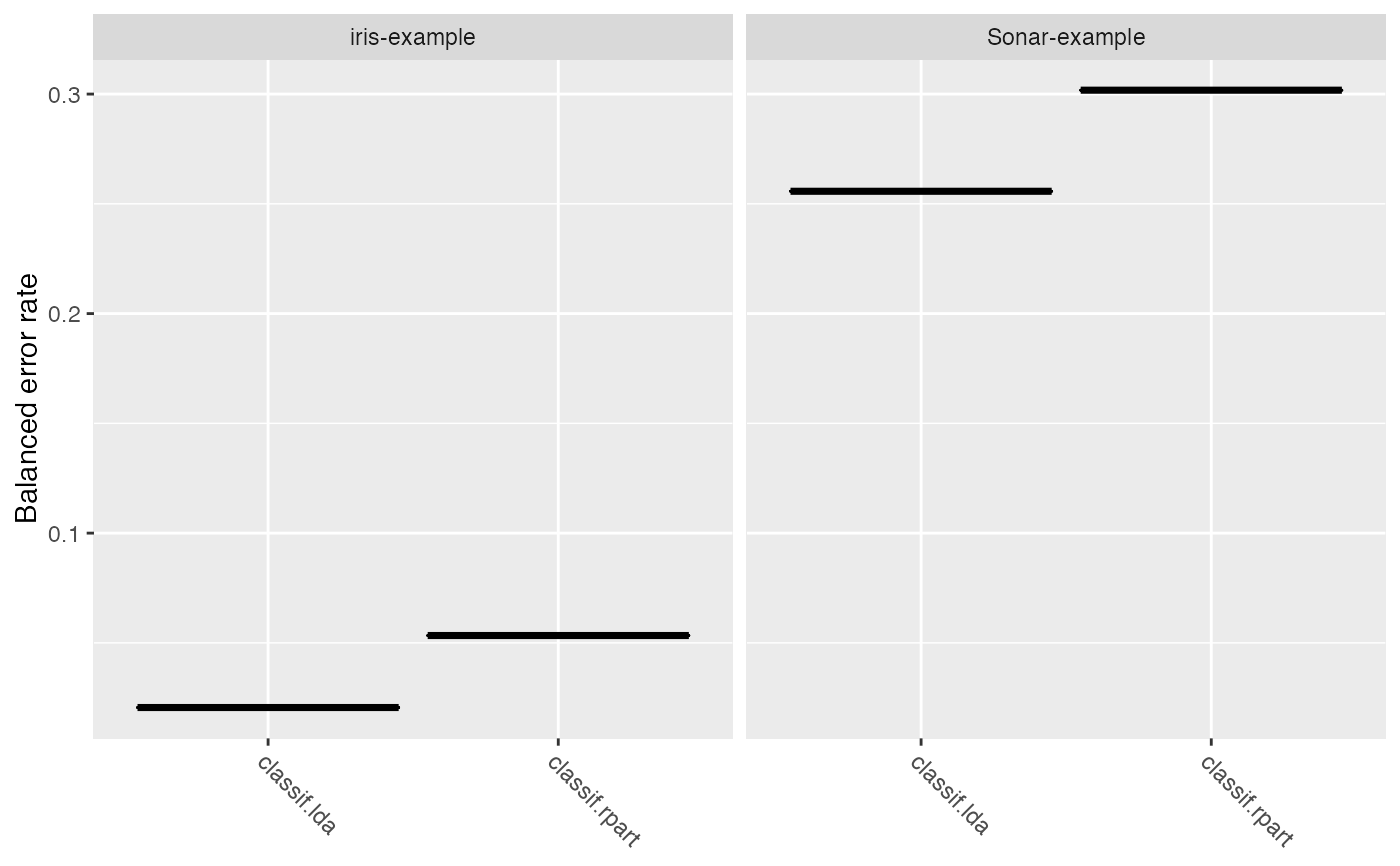
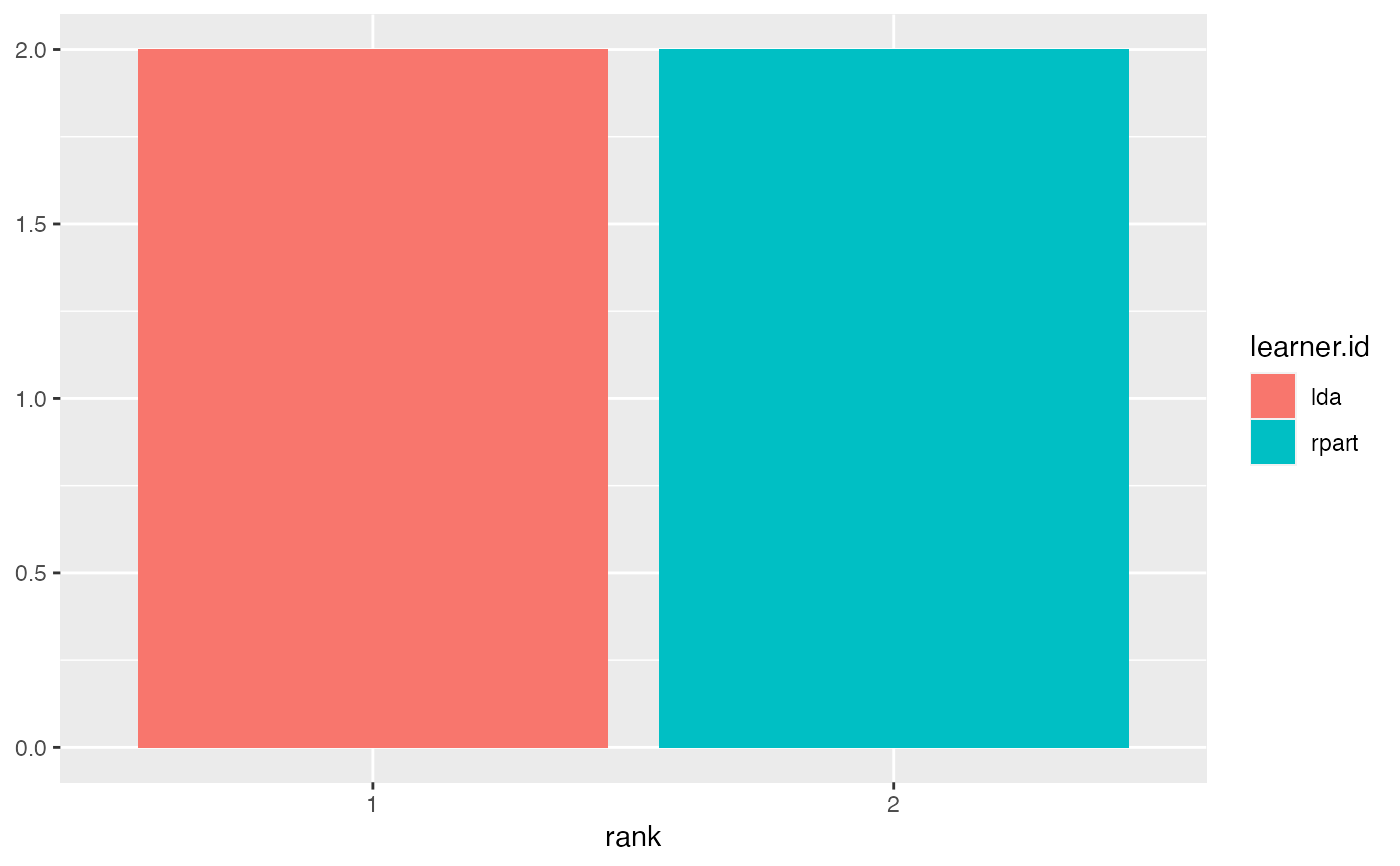
lrns = list(makeLearner("classif.lda"), makeLearner("classif.rpart")) tasks = list(iris.task, sonar.task) rdesc = makeResampleDesc("CV", iters = 2L) meas = list(acc, ber) bmr = benchmark(lrns, tasks, rdesc, measures = meas)#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#> Sonar-example iris-example #> classif.lda 1 1 #> classif.rpart 2 2#> Warning: `fun.y` is deprecated. Use `fun` instead.#> Warning: `fun.ymin` is deprecated. Use `fun.min` instead.#> Warning: `fun.ymax` is deprecated. Use `fun.max` instead.#> Warning: no non-missing arguments to max; returning -Inf#> Warning: Computation failed in `stat_ydensity()`: #> replacement has 1 row, data has 0#> Warning: no non-missing arguments to max; returning -Inf#> Warning: Computation failed in `stat_ydensity()`: #> replacement has 1 row, data has 0#> #> Friedman rank sum test #> #> data: acc.test.mean and learner.id and task.id #> Friedman chi-squared = 2, df = 1, p-value = 0.1573 #>#>#> Warning: Cannot reject null hypothesis of overall Friedman test, #> returning overall Friedman test.#> #> Friedman rank sum test #> #> data: acc.test.mean and learner.id and task.id #> Friedman chi-squared = 2, df = 1, p-value = 0.1573 #>