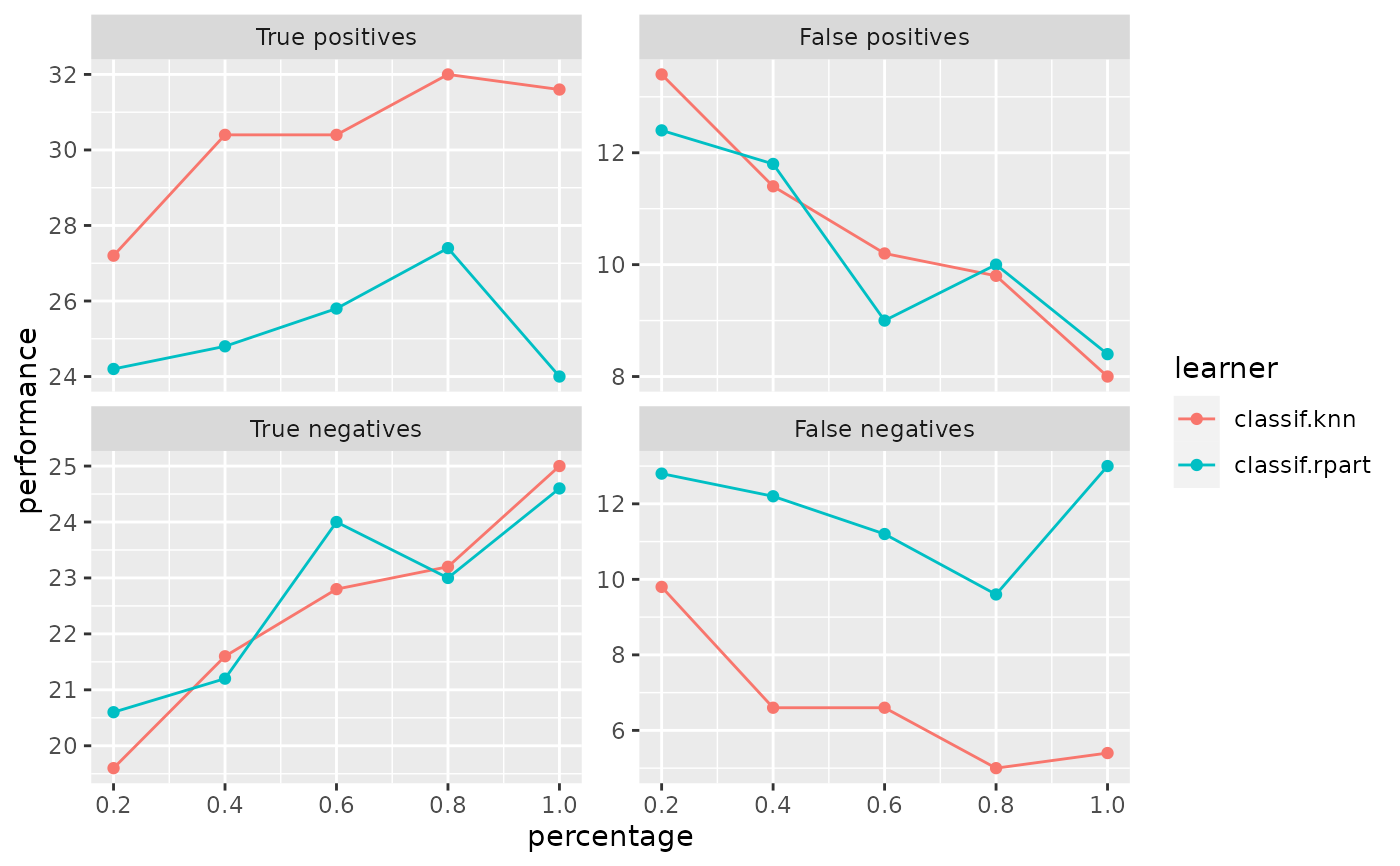
Observe how the performance changes with an increasing number of observations.
Usage
generateLearningCurveData(
learners,
task,
resampling = NULL,
percs = seq(0.1, 1, by = 0.1),
measures,
stratify = FALSE,
show.info = getMlrOption("show.info")
)Arguments
- learners
[(list of) Learner)
Learning algorithms which should be compared.- task
(Task)
The task.- resampling
(ResampleDesc | ResampleInstance)
Resampling strategy to evaluate the performance measure. If no strategy is given a default "Holdout" will be performed.- percs
(numeric)
Vector of percentages to be drawn from the training split. These values represent the x-axis. Internally makeDownsampleWrapper is used in combination with benchmark. Thus for each percentage a different set of observations is drawn resulting in noisy performance measures as the quality of the sample can differ.- measures
[(list of) Measure)
Performance measures to generate learning curves for, representing the y-axis.- stratify
(
logical(1))
Only for classification: Should the downsampled data be stratified according to the target classes?- show.info
(
logical(1))
Print verbose output on console? Default is set via configureMlr.
Value
(LearningCurveData). A list containing:
The Task
List of Measure)
Performance measuresdata (data.frame) with columns:
learnerNames of learners.percentagePercentages drawn from the training split.One column for each Measure passed to generateLearningCurveData.
See also
Other generate_plot_data:
generateCalibrationData(),
generateCritDifferencesData(),
generateFeatureImportanceData(),
generateFilterValuesData(),
generatePartialDependenceData(),
generateThreshVsPerfData(),
plotFilterValues()
Other learning_curve:
plotLearningCurve()
Examples
r = generateLearningCurveData(list("classif.rpart", "classif.knn"),
task = sonar.task, percs = seq(0.2, 1, by = 0.2),
measures = list(tp, fp, tn, fn),
resampling = makeResampleDesc(method = "Subsample", iters = 5),
show.info = FALSE)
plotLearningCurve(r)